Learning Graph Neural Networks with Deep Graph Library
Recording of the talk can be found here.
Overview of Graph Neural Networks
- Tasks in graph learning
- node classification (fraud detection)
- link prediction (eg: recsys)
- graph classification (eg: drug discovery)
- graph learning has 2 steps:
- generate low-dim embedding of node
- use standard classifiers from there onwards
- GNNs can learn node, edge, graph embeddings in an end-to-end fashion and are
based on message-passing between neighbors
- aggregation operation needs to be permutation invariant
- thereby these nets integrate node/edge/graph features as well as topology in a non-linear fashion
- for graph classification, there'll be a final "readout" layer to compute the overall graph embedding based on embeddings of each node
- training of large graphs is done via mini-batch training
- with pruning of neighborhood via sampling to reduce computational complexity
Deep Graph Library - an update
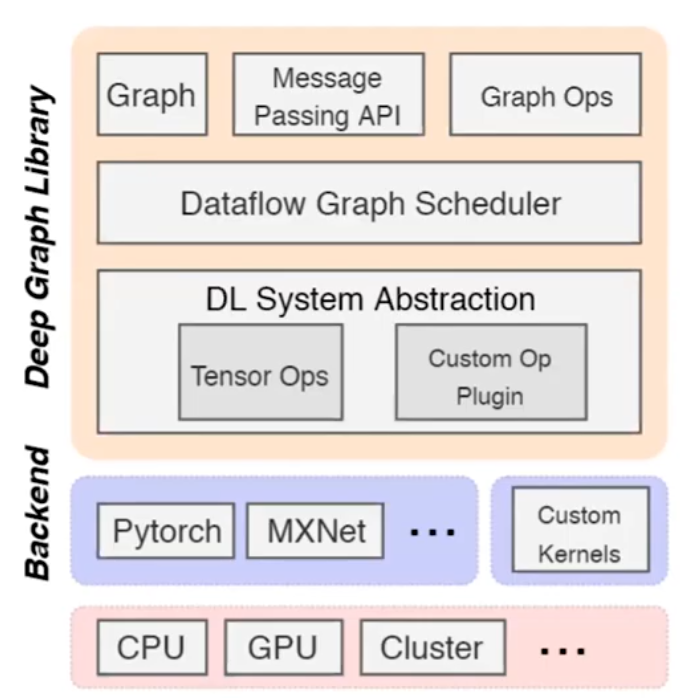
- already having active customers using DGL via AWS Sagemaker
- started out with multiple backends
flexible message propagation
- full propagation
- propagation by graph traversal: sampling on ego-network
- propagation by random walk
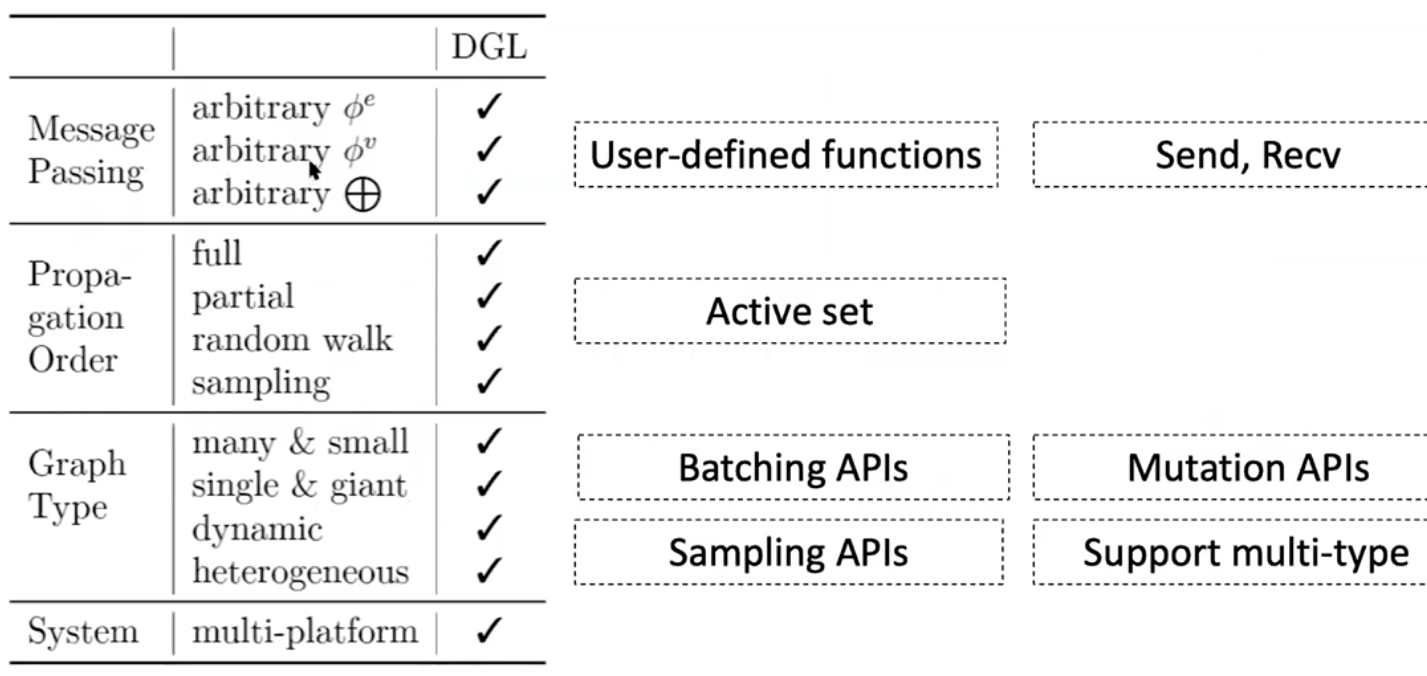
DGL programming interface
DGLGraphis the core abstractionDGLGraph.ndata["h"]- the node representation matrix
- simple and flexible message passing APIs
- active set - set of nodes/edges to trigger computation on
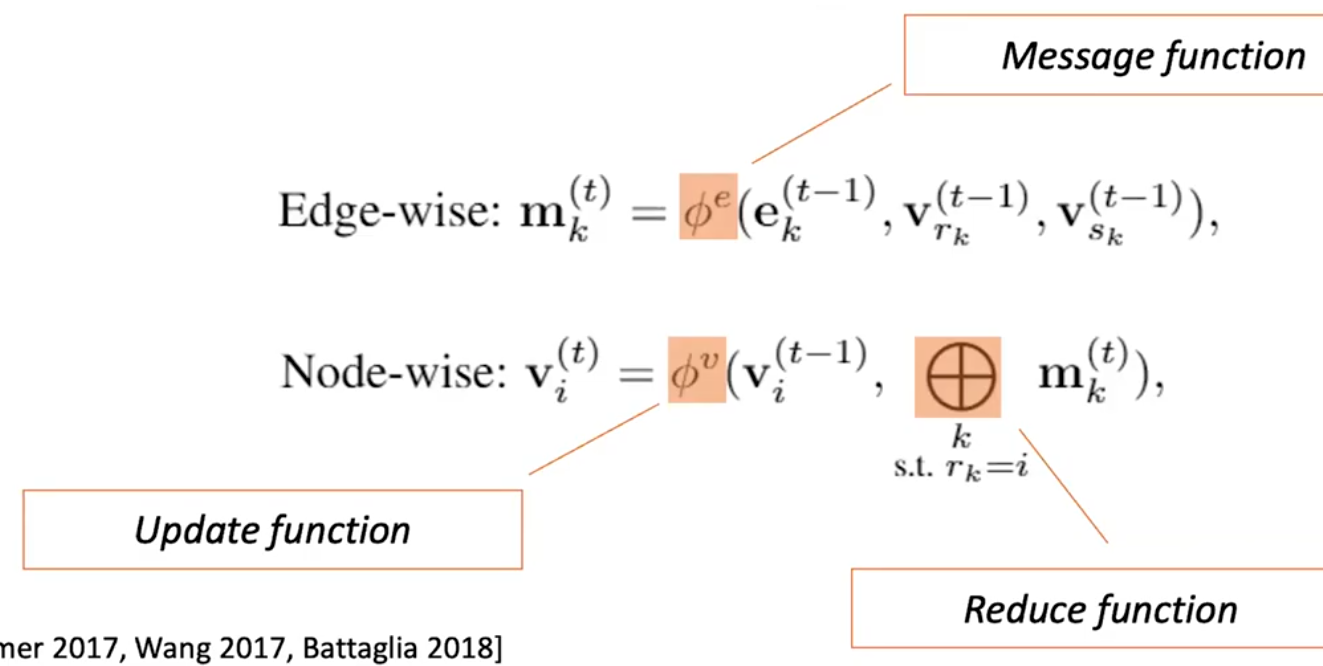
- three user defined functions
- $$\phi^v$$ - transformation function on vertices
- $$\bigoplus$$ - reduction or aggregation function
- $$\phi^e$$ - transformation function on edges
update_all- shortcut for
send(G.edges()); recv(G.nodes()); - in other words, do a full propagation
- shortcut for
- now heterogeneous graph is supported
- new sampling API is introduced in v0.43 release
- next plan is to look at distributed training
Using GNNs for basic graph tasks
- using Zachary's karate class network to demo APIs of DGL
- DGL expects node id's to be consecutive integers starting from 0
dgl.graphis the main graph structure which provides IO and query methodsdgl.graph.ndatamember is a dict that holds node features as tensordgl.graph.edatamember is a dict that holds edge features as tensor- definition of models (and their training) in dgl is similar to pytorch
How to customize graph-conv using message passing APIs
dgl.graph.ndata(andedata) can be locally updated usingdgl.graph.local_scope()- Message passing APIs in DGL are a generalization as found in
Message Passing Neural Networks. The relevant equations are as follows:
- $$m_{uv}^{(l)} = Message^{(l)}(h_u^{(l-1)}, h_v^{(l-1)}, e_{uv}^{(l)})$$
- $$m_v^{(l)} = Aggregation_{u \epsilon N(v)}(m_{uv}^{(l)})$$
- $$h_v^{(l)} = Update^{(l)}(h_v^{(l-1)}, m_v^{(l)})$$
Scale GNN to giant graphs using DGL
- for large batches it is recommended to use mini-batch training procedure
- minibatch generation on graphs
- sample the target nodes
- not done inside DGL
- using
numpy.random.choiceortorch.utils.data.DataLoader
- randomly sample the neighbors (multi-hop)
dgl.sampling.sample_neighborsfor one layerdgl.in_subgraphsimilar to the above, but will copy all edges
- construct the minibatch
dgl.to_block- renames nodes to be consecutive (for memory efficiency as well as perf)
- constructs a bipartite graph for message passing (COO format)
- sample the target nodes
to_block()has an optioninclude_dst_in_srcto help with self-loops during aggregation- inference
- we need to infer for all nodes in each layer
- thus, inference is typically costlier than training!
DGL on real world applications
- eg: recommender systems using GCMC
- introduced traditional collaborative filtering based approaches
- such a user-item matrix can be converted into bipartite graphs
apply_edges()computes edge features- heterogenous graphs:
- graphs with different types of nodes and/or edges. (eg: user-item graphs)
dgl.heterographfor creating such graphs- accessing node features is via:
g.nodes["ntype"].data["name"] - accessing edge features is via:
g.edges["etype"].data["name"]